Chemistry International
Vol. 21, No.2, March 1999
1999, Vol. 21
No. 2 (March)
.. News from IUPAC
.. Other Societies
.. Reports from IUPAC
Bodies
.. New Books
..
Reports from Commissions
.. Prizes and Awards
.. Conference Announcements
.. Conferences
CI
Homepage
News from IUPAC
Bioinformatics and the Internet
Introduction
Explosive Growth of the World Wide Web
Life Sciences and the World Wide Web
Protein Sequencing Databanks
Bioinformatics Databanks and Web Sites
Challenges to Bioinformatics
Future of Bioinformatics
ReferencesProtein Sequencing Databanks
When DNA sequence information started to grow exponentially during the 1980s, three DNA sequence databanks were established as GenBank (National Center for Biotechnology Information) in Bethesda, MD, USA; the European Molecular Biology Laboratory (EMBL/EBI) Nucleotide Sequence Database, now at the European Bioinformatics Institute (EBI) in Hinxton, UK; and the DNA Data Bank of Japan (DDBJ), Mishima, Japan, serving as mirror sites to each other.
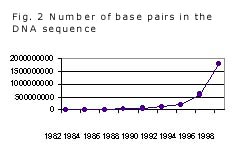
As shown in Fig. 2, the DNA databases contained 40,000 DNA sequences with a total of 50 million base pairs in 1990, but within only a decade this number has increased 40-fold, now reaching 2 billion base pairs. This increase is due largely to advances in DNA technology and robot-assisted sequencing, allowing a shift from genetics to genomics; by now, the complete genomes of 14 bacteria, baker's yeast, 12 viruses and organelles, and the nematode Caenorhabditis elegans have been published on the Internet, and many others are approaching completion, among them the human genome with a total of about 3 billion base pairs alone. This enormous increase in numbers made new types of databases possible and necessary, e.g., web sites devoted to particular organisms such as the chromosome maps of the mouse. As the number of sequenced genomes increases and can be compared to individual geno- and phenotypes ("polymorphisms"), more and more important conclusions about the structure and regulation of single genes and proteins and their interrelation in health and disease can be drawn.
On the level of individual proteins, the first sequence databanks were set up in the mid 1980s, including SwissProt at the Swiss Institute of Bioinformatics, Geneva, Switzerland, and the Protein Information Resource established by the National Biomedical Research Foundation, Washington, DC, USA. When protein structure analysis by X-ray crystallography and later by NMR spectroscopy began to grow rapidly in the 1970s, the Protein Data Bank (PDB) was established at the Brookhaven National Laboratory, Upton, Long Island, NY, USA. It contains at present over 9000 entries on protein structures. Protein science, for a long time focused on protein structure and architecture, is now in a vigorous development in its own right; comparison of protein sequences based on DNA analysis and prediction of their tertiary structure ("from sequence to structure") is an active area of research, fueled by the quest for the so-called proteome, the sum of proteins expressed by a genome under different conditions of regulation and metabolism.
Bioinformatics Databanks and Web Sites
Table 1 lists a few important examples of the many extremely useful web sites related to the life sciences. Much of the experimental work required to arrive at such findings includes the use of complex algorithms which can, in turn, often be found on appropriate Internet pages. Finally, owing to its widespread accessibility, the Internet has also become a huge blackboard for scientific information, including online versions of scientific journals, free science information (such as the public database PubMed offered over the Internet by the National Library of Medicine at Bethesda, MD, USA, which allows free access to over 9 million scientific publications), tutorials, conference announcements, and information on grants and job offers. As a particular consequence of the Internet, the access to information of scientists working in less developed countries has dramatically increased. Thus, as just four among dozens of examples, there now exist the following web sites:
http://www.sci-ctr.edu.sg/apnstc/
http://www.healthnet.org/afronets/enhr.htm
http://www.yorku.ca/research/crs/prevent/warn.htm
http://www.idrc.ca/acacia/outputs/op-unin.htmChallenges to Bioinformatics
The present shift from sequencing single genes to sequencing whole genomes is expected to expand widely our understanding of the regulation of expression, the interaction of proteins, and, finally, of the function of cells and multicellular organisms. Such progress implies new challenges to bioinformatics. There are at present two major problems:
As a probable consequence of all these developments, the biological and biochemical experiments of the future will, to some extent, be carried out not only in vivo and in vitro, but also in silico. Biology-related information will be the pertinent raw material, available from databases through the WWW, which can be profitable. As seen already in the case of the "gene hunt in silico", it becomes more and more feasible to transform this computer-based information into valuable research results or even products. Thus, it is becoming a reality that novel targets for drugs or new powerful biocatalysts can be identified in the huge and growing mass of computer-based genomic sequence information and that metabolic fluxes in living beings can be clustered, via a bioinformatics approach, to allow the genetic reengineering of metabolic pathways in microorganisms, plants, animals, or man.
- Internet Domain Survey, July 1998, http://www.nw.com/zone/WWW/top.html;
The Netcraft Web Server Survey,
http://www.netcraft.com/Survey/;
Internet Statistics: Growth and Usage of the Web and the Internet, http://www.mit.edu/people/mkgray/net/;
eMarketer,
http://www.e-land.com/;
Hermes project,
http://www-personal.umich.edu/~sgupta/hermes/- The Internet2 project,
http://www.internet2.edu/
News and Notices - Organizations and People - Standing Committees
Divisions - Projects - Reports - Publications - Symposia - AMP - Links
Page last modified 11 June, 1999.
Copyright © 1997, 98, 99 International Union of Pure and Applied Chemistry.Questions or comments about IUPAC, please contact the Secretariat.
Questions regarding the website, please contact [email protected]