BP03 From the primary to the quaternary structure
Aim: To explain the four important levels in the construction of a 3-dimensional protein structure using haemoglobin as the example. |
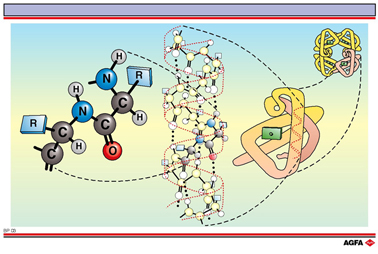
A protein has a complex three-dimensional structure that is built up in four levels. Three of these (primary, secondary and tertiary structure) are common whilst the fourth level (quaternary structure) requires the protein to contain more than one polypeptide chain.
Primary
structure
Proteins are polymers with a specific number and order of amino acids.
This order, passed on by the genetic code, is called the primary structure
and is the basis for the further folding of the protein chain.
Secondary
structure
A protein chain is not a linear molecule: locally the chains fold themselves in a specific
manner. The alpha helix is an example of this regular folding. Hydrogen bonds play an
important role in the stabilisation of this secondary structure.
Tertiary structure
Helix domains can in turn fold to give a complex structure.
In myoglobin eight alpha helix domains are folded around a central heme group (coloured
green on the illustration). Chiefly non-covalent interactions such as electrostatic
forces, hydrogen bonding, and dipole-dipole interactions contribute to the stability of
the tertiary structure.
Quaternary structure
Proteins can sometimes exist in the cells as aggregates of two or more folded polypeptide
chains called sub-units. Haemoglobin is for example built up of four folded chains, each
of which has a structure analogous to that of myoglobin.
The prediction of the three-dimensional structures of proteins, via computer simulation based on the primary structure, is still a dream for most proteins, although it is usually possible to do this for the secondary structure. When the protein can be crystallised however it is possible to determine the three-dimensional structure experimentally using X-ray diffraction. An important part of our knowledge of the three-dimensional structure and the structure-activity relationships of proteins is based on this technique.